Plastic Circular Economy
Australia has a plastic disposal problem…
Since Chinese firms stopped burning our waste in exchange for cash, the true scope of Australia’s plastic waste crisis has been revealed. We don’t have a plan and recycling firms lack the technological and financial investment to do it on their own.
How can Synthetic Biology solve the plastic crisis?
Plastic polymers are incredibly difficult to break down chemically, which is why we’ve so often resorted to burning plastic landfills rather than reusing. Enzymes are biological catalysts that can rapidly increase the rate of reactions on a molecular scale. Plastic in the environment is a potential energy & carbon source - and evolution never lets a niche exist unoccupied for long. Every day that our environment is saturated with plastic is another opportunity for an evolutionary event to occur that allows a microorganism to digest that plastic, faster, more efficiently. Without intent, simply through trial and error, these microorganisms take advantage of this latent resource by slowly constructing an enzyme fit for the purpose of digesting it. Careful research and observation of plastics in landfill that degrade especially rapidly will result in the discovery of plenty of enzymes that could be used industrially. Once discovered, we can insert the gene for producing the enzyme of interest into our lab bacteria and produce large quantities for industrial use. The real challenge is searching out and finding the right enzymes.
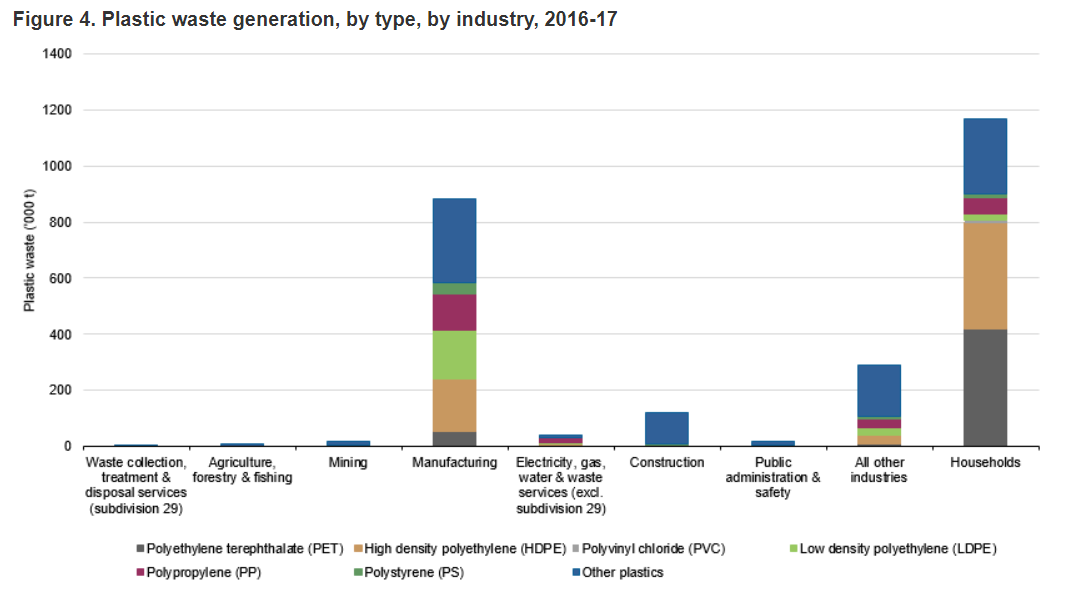
What plastic needs breaking down?
According to the Australian Bureau of Statistics, a large majority of plastic waste consists of just two polymers:
Polyethylene terephthalate (PET), 419,383 tonnes (35.9%)
High density polyethylene (HDPE), 337,544 tonnes (32.3%)
If we could magically break down 70% of our domestic plastic waste with just two enzyme systems, that’d revolutionise the industry. It’d be great to deal with LDPE and the ominously named ‘other plastics’ category as well… and we will. But let’s walk before we run.
So what enzyme systems do we need to break down PET and HDPE?
PET
PETase was the first enzyme that truly caught my eye, and there is a ton of excellent research on it - but I eventually came to the conclusion that it was too inefficient for too industrial use. Then came this paper:
An engineered PET depolymerase to break down and recycle plastic bottles
PET Depolymerase is an entirely different story, providing orders of magnitude more efficiency than PETase. To quote the French inventors (V. Tournier, C. M. Topham, A. Gilles, B. David, C. Folgoas, E. Moya-Leclair, E. Kamionka, M.-L. Desrousseaux, H. Texier, S. Gavalda, M. Cot, E. Guémard, M. Dalibey, J. Nomme, G. Cioci, S. Barbe, M. Chateau, I. André, S. Duquesne & A. Marty) that authored the paper with this enzyme’s sequence;
“Here we describe an improved PET hydrolase that ultimately achieves, over 10 hours, a minimum of 90 per cent PET depolymerization into monomers”
The authors managed to digest a full ton of plastic bottles down to monomers within this 10 hour period. Truly remarkable, and thanks to their publication - anyone can reverse engineer their protein and try to replicate these results! So I did just that…
We can then codon optimise these for E. Coli expression, converting the Amino Acid code to Nucleic Acid Code. I chose to do this for the most efficient enzyme variant discovered by the paper.
We can then take this gene and insert it into an E. coli plasmid fit for large scale protein expression. I chose the plasmid pUS24X with it’s cumate inducible expression system for the job:
Unfortunately, this gene comes with a leftover expression tag. This is half of an unknown system, since the original enzyme was discovered by metagenomic analysis of a compost heap, rather than of a specific organism. Since we can’t find the other half of the expression system - I propose that we eventually try removing it to see if there is a negative effect on the efficiency of the system. For now, it’s best not to interfere with something that works - so I decided to include this random signal peptide alongside the enzyme itself.
HDPE
HDPE is yet to have an exciting breakthrough enzyme like PET depolymerase, but that doesn’t mean it doesn’t exist. Nature continues to prototype millions of enzymes every day and it just takes one observant researcher to discover the next great molecular machine.
Promising fungal research have turned up potential candidates, but these suffer from the lack of efficiency we observed in PETase. Nonetheless, here are some links to interesting papers on the topic;
Expect this page to be updated as soon as an excellent HDPE degrading enzyme is discovered!
What’s left after the Enzymatic Digest?
Plastics are generally simple polymers; effectively long chains of 1-2 molecules, repeating ad infinitum. Thus, when you break down a large piece of plastic into its individual components you’ll eventually end up with monomers - the individual molecules in the chain.
These aren’t necessarily better than macroplastics and the microplastics produces along the way are especially worrying in their impact on the environment. Thus the ideal way to digest plastics is in a bioreactor where the monomers can be collected and stored for downstream use.
PET Monomers
Terephthalic acid
Ethylene glycol
HDPE Monomer
Ethylene
How do we take advantage of the plastic monomers produced?
What’s better than removing tonnes of plastic from the global ecosystem and reducing the burden on local landfills? Making a ton of money while doing so.
The monomers can be purified and reused to produce recycled plastic, however we’re entering a world where biomass is becoming a critical resource. Industries ranging from pharmaceutical to textile will increasingly begin to demand biomass as their supply chain begin to rely on it as a primary input.
If we can find strains of microorganism that rapidly grow on plastic monomers, using them as a Carbon or Energy source - we can close the circle and create a profitable Circular Economy.
List of Bacteria that can use these monomers as a Carbon or Energy Source.
Terephthalic acid
Rhodococcus sp. strain DK17 Comamonas testosteroni YZW-D [49], and Comamonas sp. strain E6
Ethylene glycol
Flavobacterium
Acetobacterium woodii
Ethylene
Is a plant hormone, problematic to release large amounts of it in the environment.