Alex Kelly
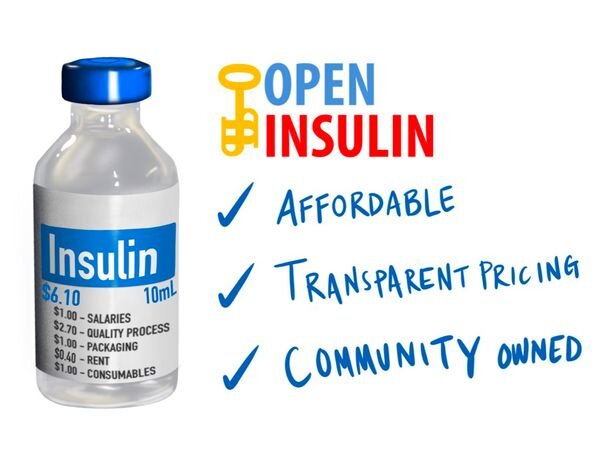
Open Insulin australia
Open Insulin was started in California, USA by Counter Culture Labs as a response to the price of insulin jumping 300% in a decade, despite it being an off-patent medication. This price rise defied logic, so they theorised a logic-defying initiative would be the solution. They set out to make every step of the insulin production process Open Source.
When I joined as a collaborator in 2016, their project became an international one. The challenge was no longer to break down systemic issues within American medical supply chains, but one of purchasing power, wealth inequality, international distribution and segmented regulatory jurisdiction. Collaborators from every continent (except Antarctica) now regularly tune in to our global call and we seek to address this challenge as a united humanity, rather than nationals clamouring in a zero-sum supply game.
2016
Halfway through my Commerce/Science degree, I asked to join Open Insulin as an international collaborator. Since Insulin is more affordable in Australia thanks to healthcare subsidisation, I focused on the international problem instead.
Team: Alex Kelly
Host Lab: Biofoundry
Highlights:
Open Insulin became an international collaboration this year.
While planning and researching I met Dr. Nicholas Coleman of the University of Sydney (USYD), who took an interest in the project.
Challenges:
I was still a novice in the lab and to understand protein expression I needed to read scientific papers for 6 months. By the end I’d accumulated over 100 papers which I organised by topic - a resource that would come in handy next year!
2017
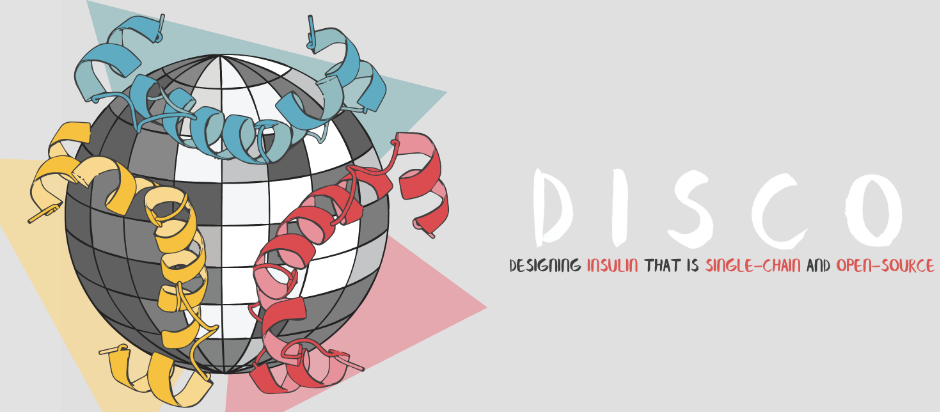
The year that everything changed. My project was selected by Dr. Coleman to be USYD’s submission to iGEM 2017 (The International Genetically Engineered Machine). This competition is the largest gathering of Synthetic Biologists under one roof and is a huge deal! The DISCO project (Designing Insulin to be Single-Chained and Open-Source) was born!
Team: Dr. Nicholas Coleman, Emaleen Najjar (Team Captain, aka. Boss Lady), Courtney Pratt, Erica Stewart, Grace Attrill, Jamie Littleboy, Ruby Dawes, Stephanie Sun, Alex Kelly
Host Lab: Coleman Labs, USYD
Highlights:
THIS WIKI
We designed a novel insulin analogue and expressed it (alongside human proinsulin) in E. coli bacteria.
We successfully improved the yield of correctly folded protein using an Ecotin fusion tag for periplasmic expression. We travelled to the iGEM Jamboree in Boston to show off our work!
After the Jamboree, I travelled to California to meet the American team in person for the first time. I also got to visit the Counter Culture Labs space.
Challenges:
The learning curve was so steep this year that I chose to drop out of my university courses at UNSW for the semester in order to fully focus on Open Insulin.
We were unable to model purification using an FPLC due to a lack of budget or equipment. Instead we resorted to more manual methods, using a centrifuge. This made the modelling of pricing more difficult.
2018
Now lets do it all again, but with no budget and broken equipment.
Team: Alex Kelly, Andrew Kirkby, Benj Gonzaga, Sarra Mrad, Dani Henry Forster, Dani Fox, Daniel Gilbert, James Newman, Mason Nicholas
Host Lab: Biofoundry
Highlights:
I traveled to Geneva for the 2018 Geneva Health Forum where I was invited to present a stall about Open Insulin for 3 days, making many new friends.
We began work on developing low-cost protocols, experiencing the satisfaction of replacing a $200 reagent with something we mixed for less than $1.
Challenges:
Everything is harder and it was rare that we had a day where everything went right. Yet through adversity, came camaraderie.
2019
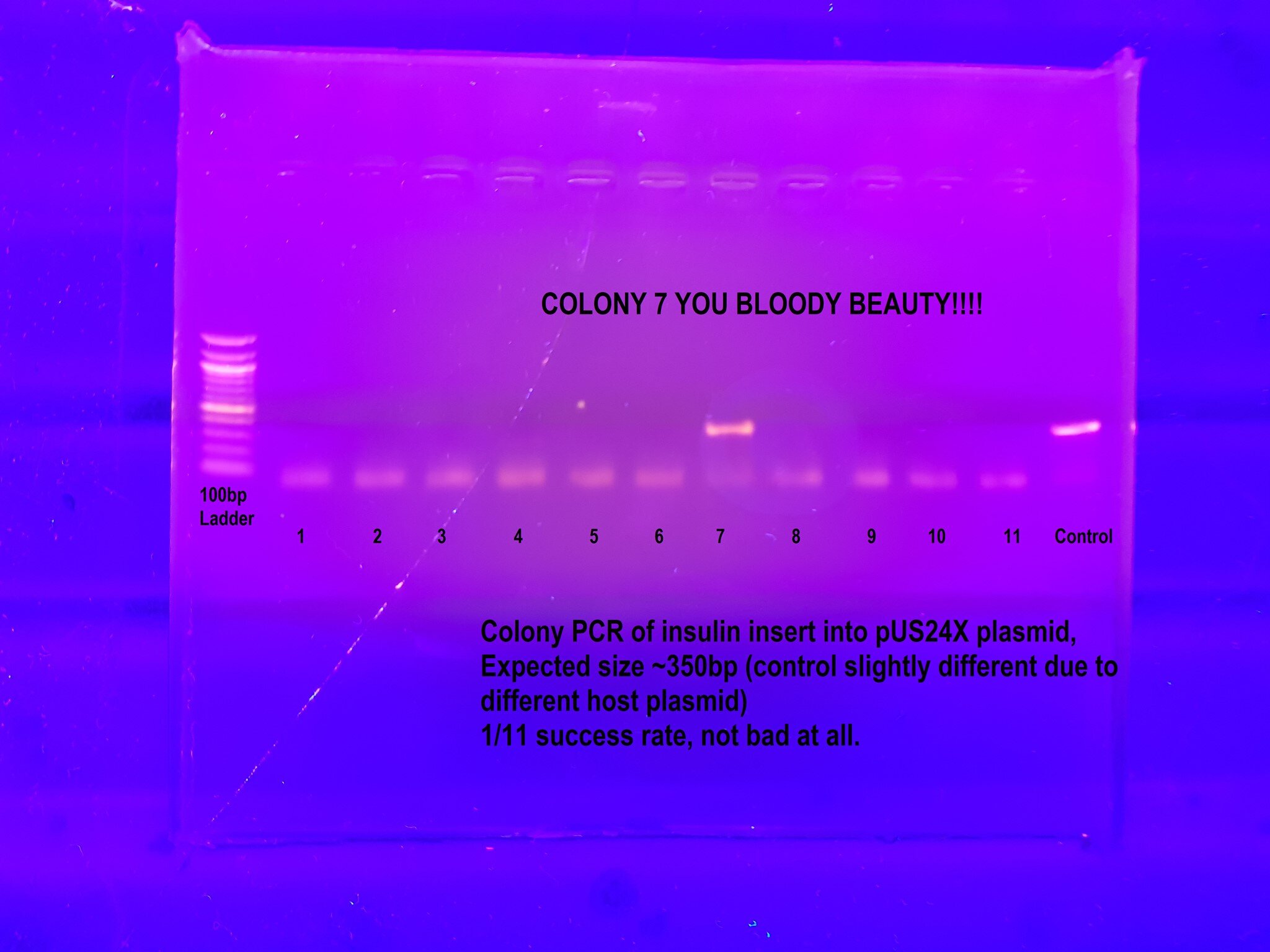
Pushing through adversity, we advanced the project a single step! After 18 months of fixing equipment and mixing reagents, we successfully cloned our gene into a new backbone - pUS24X then verified it via PCR. It was a small step forward from the giant leaps and bounds of 2017, but we were proud to have done it with a shoestring operating budget.
Team: Team: Alex Kelly, Benj Gonzaga, Andrew Kirkby, Oliver Dibley, Sarra Mrad, Henry Forster, Danielle Fox, James Newman, Mason Nicholas, Shaun Shankar, Soroush JP, Jeremy Thomas, Adam Carmichael, Alan Li
Host Lab: Biofoundry
Highlights:
I was invited to speak at the 2019 Experts on Diabetes Summit in Geneva, an initiative run by the Geneva Health Forum in collaboration with the World Health Organisation
Dr. Gojka Roglic of the WHO praised my concept of an international repository of biosynthetic bacteria.
The WHO would then go on to announce that it would start accrediting insulin for the global market, expanding the market gatekeepers from the original two (FDA + EMA) to a total of three. This could be a change with massive implications, but only time will tell...
Back in the lab: Using an open source plasmid provided from Coleman Labs superstar Mark Somerville, we successfully upgraded the expression system of our Cytoplasmic Proinsulin gene. T7 Expression System >> Cumate Operon.
Challenges:
Equipment and resource challenges meant that we haven’t yet done the same for the remaining 3 genes (PPro, CWin, PWin).
I still need to confirm the insert via flanking PCR and sequencing.
2020/2021
Unfortunately due to the pandemic, I’ve been unable to get lab access to continue the project.
However! 2021 is the centenary of the discovery of Insulin by Banting, Best and Collip. Unable to work in the lab to push the project forward, I wrote the following to celebrate it;
Biohacking: Potential for Disruption
Despite insulin’s role as the model protein for technological innovation, much of the population of people with diabetes in low- and middle-income countries have challenges accessing this essential medicine. Unlike chemically synthesised medicines, the expiry of patents has not resulted in a traditional patent cliff - whereby the price rapidly drops to a new equilibrium as competitors enter the market. Paradoxically, the price of insulin has instead risen as patents expire and the technologies that were pioneered with insulin become available for use by anyone. This lack of competition can most likely be attributed to the extraordinary barriers to entry that exist within the biomedical market.
An unlikely revolution stands poised to challenge this status quo, as the field of biohacking seeks to disrupt the extant oligarchic biosimilar market structures via the mainstream dissemination of innovation and manufacturing protocols. Echoing the impact of the personal computing revolution on the computer science industry, biohackers (aka. DIY biologists) make incremental improvements to well characterised biological systems. Connected internationally through online communities, these independent researchers have formed a modern day Respublica literaria that allows for an unprecedented sharing of knowledge that subverts the traditional publishing industry.
The greatest challenge historically faced by prospective insulin manufacturers is a lack of vertical standardisation in the approval process. A complete accounting of every variable within a living biological system is extremely difficult, even if the process is copied exactly from an expired patent. As a result, despite the best efforts of the FDA and EMA to streamline the process, only two ‘biosimilar’ insulin products have thus far made it through biosimilar clinical trials in the USA. Neither is considered interchangeable with its generic counterpart, further complicating their usage for diabetic patients. In 2019 the World Health Organisation volunteered to help by accrediting additional generic insulins, however they face the same non-standardisation hurdle as the existing regulators.
A solution for safely standardising the top-bottom synthesis of biosimilars must be found before regulators will feel comfortable unleashing a sufficient pipeline of generics into the market. This standardisation of biosynthesis requires a paradigm shift in the approaches and methodology of molecular biology. The emergent field of synthetic biology, exemplified by the iGEM competition, offers a practical demonstration of what this shift will look like. Through the application of an engineering philosophy (Design > Build > Test > Learn) alongside rapid iterative cycles, researchers can accurately elucidate the impact of individual changes in input variables on the finished insulin product. The abstraction of genetic parts into discrete units offers an opportunity to build significantly more efficient systems for the production of insulin without compromising on quality.
Incremental innovations within this field are discovered, analysed in excruciating detail and published in an open-source manner. This approach offers the missing link in standardisation and system comprehension that biosimilar regulators desperately need in order to approve improvements in biologic manufacturing – while simultaneously subverting the potential for exclusionary patents by publishing in the public record. Codon optimisation, improved ribosome binding sites and cheaper Car9 purification tags (to replace the historically used His-tag system) are just some of the innovations that would allow a new market entrant to easily compete on price with the entrenched market occupants. History teaches that the introduction of such innovations would require years of validation and millions in regulatory approval expenses. However, the wealth of data provided by these innovative communities may begin to break down these barriers to entry.
The open-source publishing encouraged by competitions such as iGEM is one of the major reasons for the existence of the biohacking community. The sheer accessibility of the wiki format means that anyone can reverse engineer the published results, taking advantage of increasingly affordable DNA synthesis in order to contribute even more research data and innovation to the sphere of public knowledge. While such activities may currently lack the investment capital to meaningfully impact the global insulin supply, they represent a genuine grassroots disruption to a complacent market. It is inevitable that the nascent opportunities uncovered by these unaffiliated operators will be subsumed by a larger existing player in the generic marketplace. An opportunity exists here to undercut the extraordinary profit margins in the insulin marketplace and seize a sizeable market share with a more competitive product.
The burgeoning popularity of ‘biology as a hobby’ indicates a bright future for biohacking. As synthetic biology matures as a field, the alumni of competitions such as iGEM contribute to a growing skill and knowledge pool. Members of this community may be as influential on the biotechnological industry as freelance coders are on the computer science industry, or retail investors on Wall Street. With sufficient direction and organisation insulin could be the first biosimilar protein to outcompete its previous iterations with publicly available open-source technologies, rather than inventive steps defended by exclusionary patent architecture.